Two new PhD positions in computational evolution are available in the Centre for Computational Evolution at the University of Auckland to work on developing Bayesian integrative models of evolution that use data from genomic sequences, phenotypic data and the fossil record. The research will include the design and development of new mathematical and computational models for Bayesian phylogenetic inference. The successful candidates will work with an international team of computational biologists, evolutionary biologists and palaeontologists to both develop new methods and test them on a number of exciting data sets.
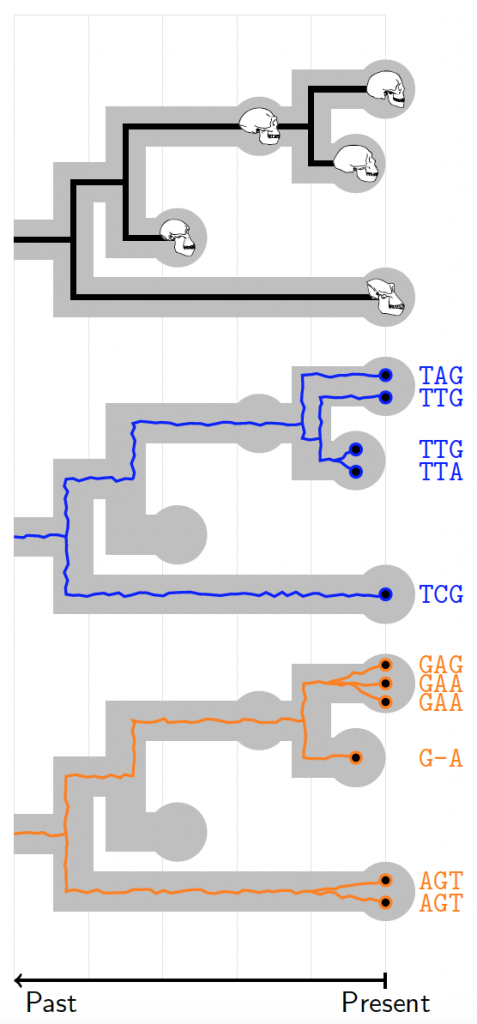
How and when species came to be is the fundamental question in macroevolution. Attempts to answer it use a variety of data sources including genome sequences, morphology and fossil discoveries. Yet current methods are unable to exploit all this data, with different data sources often producing conflicting results. This project aims to create a unifying probabilistic framework that combines genomic, fossil and phenotypic data to give us the best possible understanding of evolutionary history. The PhD research will involve creating open-source software tools to disseminate new methods widely as well as using these new methods to address outstanding questions in human, animal and pathogen evolution.
The initial focus of the work will be to extend the StarBEAST2 package to allow for sampled ancestral species and their phenotypes in the species tree as well as ancient DNA samples in the embedded gene trees. This is a major software engineering task. There will also be work on developing new trait evolution models that can account for trait variation both within and between species. Current models will be incorporated into the BEAST 2 software and major studies on real and simulated data will be run to assess their strengths and weaknesses. The successful candidates will also have the opportunity to develop new inference methods for continuous trait evolution. Finally a third focus of the work will be on incorporating rich fossil data into the phylogenetic framework within BEAST 2. New models will incorporate variability of sampling over time and space, trait-dependent sampling, and will be able to use multiple fossils from the same morphospecies while accounting for uncertainty in the geologically-derived age of fossils. Both simulated and curated data sets will be used to test and prove the newly developed methods.
The successful candidates will work with an international team including Professor Alexei Drummond, Dr David Welch (University of Auckland), A/Prof Tanja Stadler (ETH Zurich) and Dr Nick Matzke (ANU), as well as expert collaborators with knowledge of specific paleontological and molecular data sets including Dr Mana Dembo (hominins; Simon Fraser University) and Dr Graham Slater (canids; University of Chicago).
Each position comes with a stipend of $NZ27,300 (which is annually adjusted for inflation) and payment of enrolment fees. There is no teaching requirement associated with the stipend.
The successful applicants will have a strong background in a quantitative subject (such as Computational Biology, Mathematics, Statistics, Computer Science, Physics or similar), an understanding of Bayesian statistics, some experience of coding and ideally have had some exposure to, or at least a strong interest in, phylogenetic methods. The exact nature of the work will depend on the strengths and background of the successful candidates.