23 June 2015 by Remco Bouckaert
“What is going wrong?” is an often asked question. There can be many things going wrong, but there is one thing that goes wrong more often than other things and it easy to fix.
The first thing you want to check is the settings of the rates in BEAUti. There are two places where rates are set:
- The site model panel, where the substitution rate is set
- The clock model panel where the clock rate is set
The final rate used is the product of these rates.
The way to think of the substitution rate is that it is a relative rate with respect to other partitions, while the clock rate is the overall rate for substitutions per site per year (or any other unit of time you choose to use). So, substitution rates will be numbers close to 1, while clock rates tend to be small numbers, such as 4e-9 substitutions per site per year.
Substitution rates
To set up the substitution rates, use this chart:
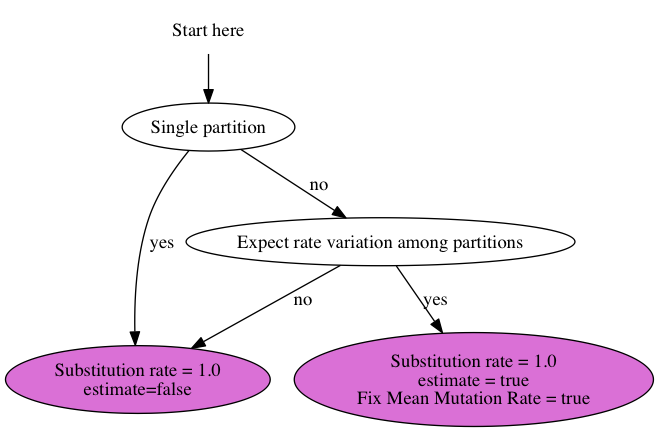
Standard analysis
For an analysis using the Standard template, you can go to the clock model tab and use this chart to set up the clock rate:
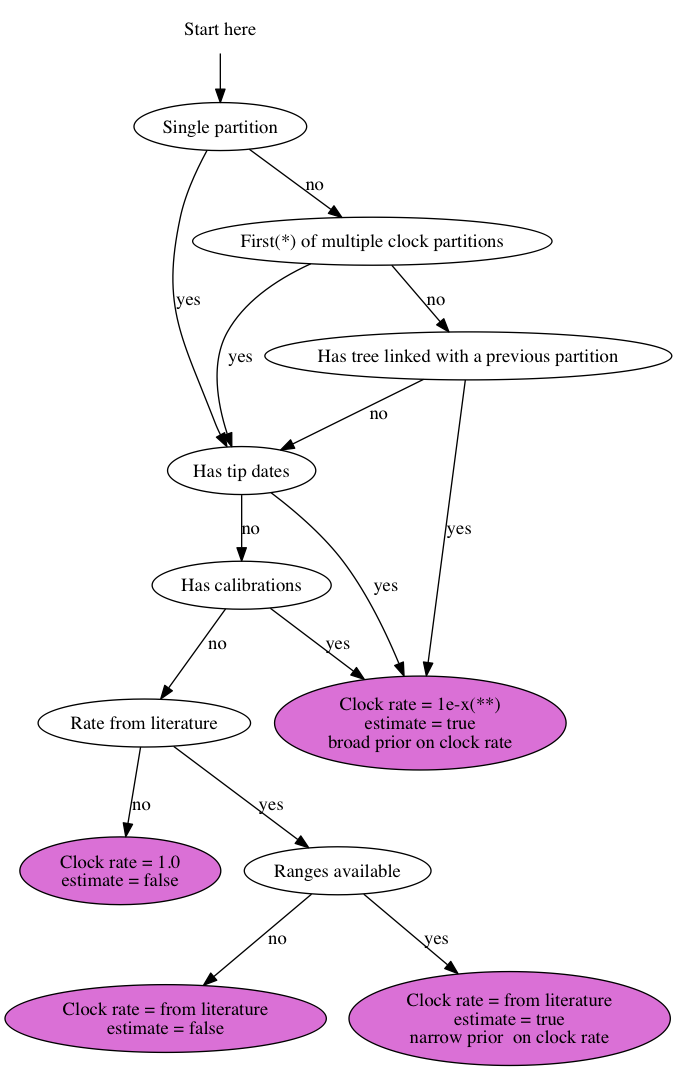
* Partitions can be ordered arbitrarily. With the first partition I mean the one for which there are either calibrations, tip dates or a rate from the literature, which usually is the first partition listed in the list of clocks, but may be a later one as well.
** Set the clock rate to “1e-x” where x is a number that is somewhere in the region you expect it for your data helps to get through burn-in faster. You could leave it at the default value of 1.0, but it just takes longer to reach convergence. Assuming you are using years as units of time, workable values are 1e-9 for nuclear data, 1e-6 for mitochondrial, bacterial and DNA viral data and 1e-4 for RNA viral data, but if you have more specific information about your sequences it helps to use it to specify starting value.
*BEAST analysis
*BEAST analysis are a bit different in that tip dates are not allowed (at the time of writing) and calibrations are on the species tree, not the gene tree. Usually, all clock rates but the first are estimated using a broad prior. To decide whether the first rate should be estimated or not, use the chart above.
If BEAST still acts weird after rates are set up correctly, just post a question on the BEAST user list.
