23 June 2014 by Remco Bouckaert
The birth-death skyline model cite{Stadler} is a tree prior that can deal with serially sampled tip dates. It can be implemented as a lean CalculationNode, which takes a set of five parameters that can be estimated as input, plus an input for the number of intervals in the skyline model. The birth and death rate, and the sampling rate have gamma priors on them, and there are a few functions of the parameters we want to log. The complete model is illustrated in below. The parameters, tree, operators, priors and loggers are already available in BEAST. The loggers use the RPNcalculator class to calculate a simple arithmetic function of the parameters. This only leaves code to be developed for the details of the birth-death skyline model itself.
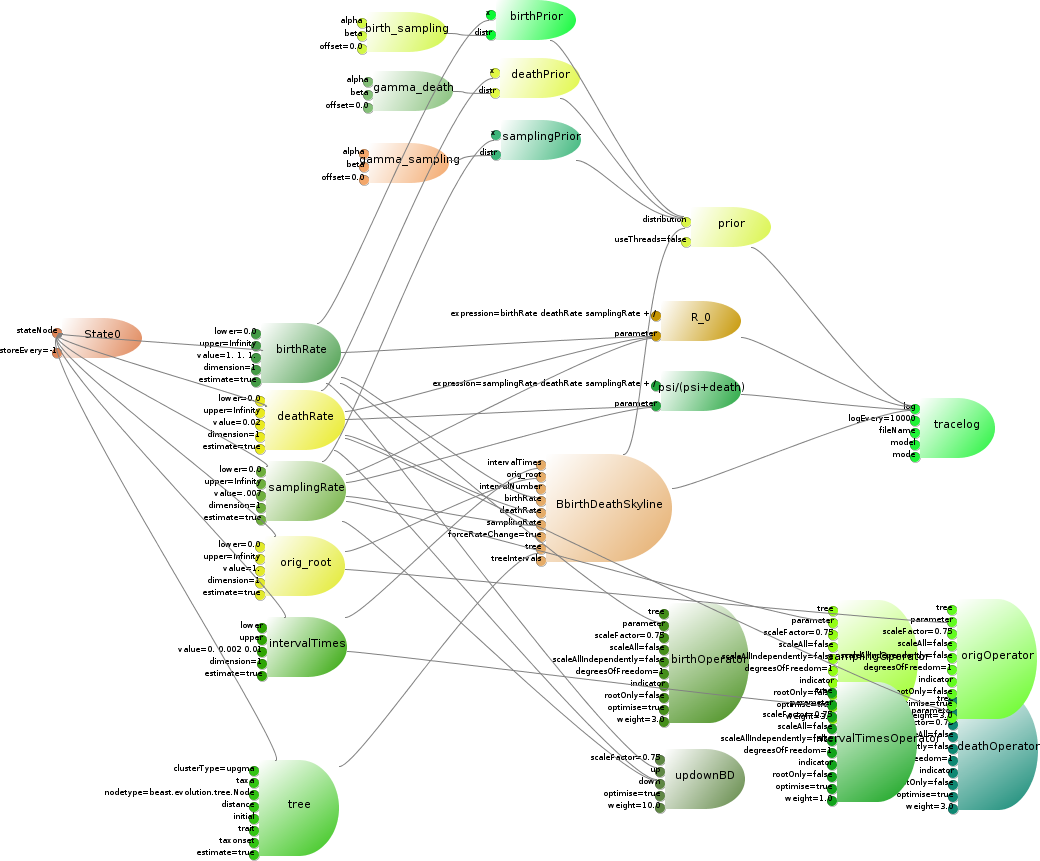
The Birth-Death Skyline model consists of a tree prior, six parameters, a number of operators on these parameters, some priors on these parameters, two specialised loggers, and a set of rules to connect model, e.g., the tree to the prior, the loggers to the trace-log, etc. For clarity, the links between parameters and state are omitted as well as the links between parameters and trace-log.
Step 1: Write BEASTObject – develop code
The code was developed by Denise K”uhnert, and we will have a look at some of the more interesting areas. It starts with a proper description.
@Description("Adaptation of Tanja Stadler's BirthDeathSamplingModel, to allow for birth and death rates to change at times t_i")
The class definition starts defining the five parameter inputs and the input for the number of intervals. Further, there is a boolean input that influences the behaviour.
public class BirthDeathSkylineModel extends SpeciesTreeDistribution {
public Input intervalTimes =
new Input("intervalTimes", "The times t_i specifying when rate changes can occur", (RealParameter) null);
...
public Input forceRateChange =
new Input("forceRateChange", "If there is more than one interval and we estimate the time of rate change, do we enforce it to be within the tree interval? Default true", true);
Then follow the member variables for shadowing input parameters and storing intermediate results. This is a thin CalculationNode so there are no member variables for storing/restoring intermediate results.
double t_root;
int m;
protected double[] p0_iMinus1;
...
}
The initAndValidate method sets up member variables and does some sanity checks on the inputs.
@Override
public void initAndValidate() throws Exception {
super.initAndValidate();
m = intervalNumber.get().getValue();
if (intervalTimes.get() != null){
if (intervalTimes.get().getDimension() != m)
throw new RuntimeException("Length of intervalTimes parameter should equal to intervalNumber (" + m + ")");
timesFromXML = true;
}
}
Then, a few hundreds of lines of code are omitted, which perform the actual calculations. The method of interest for a tree-prior is calculateTreeLogLikelihood for a tree. We refer to the actual code for details, but this is where the most interesting part of the class resides.
@Override
public double calculateTreeLogLikelihood(Tree tree) {
m = intervalNumber.get().getValue();
// details omitted ...
return logP;
}
Finally, the implementation of the CalculationNode is minimal, since this is a thin CalculationNode. The methods store and restore are not even overridden, and just use the base implementation, while requiresRecalculation is the most minimal implementation possible.
@Override
protected boolean requiresRecalculation() {
return true;
}
Step 2: Test BEASTObject
To test the new BEASTObject, write JUnit tests and create an example XML file, which typically goes in the examples directory. This example file can work as a blue-print for the BEAUti template to a large extend.
Step 3: Write BEAUti template}
We will go through the BEAUti template highlighting some of the aspects. Some of the detail is omitted for brevity and replaced by `…’ . Since a BEAUti template is in BEAST XML format, we start with a BEAST 2 header. For brevity, we introduce
This prior is a tree-prior, and wherever the main template has a merge point for tree priors, this prior should be made available. mergewith is a reserved XML element name, so no spec attribute is necessary.
This is the start of the sub-template definition. Again, there is no spec attribute, but this time it is assumed that the main template has a map-element that maps the subtemplate element name to the BeautiSubTemplate class. Likewise, connect is mapped to BeautiConnector to make things more readable.
The sub-template has an id, which is used in combo-boxes in BEAUti to identify this model. The class attribute indicate that the sub-template produces a BEASTObject of this type. One task of BEAUti is to create new BEASTObjects, and connect them to an input. The way BEAUti finds potential BEASTObjects to connect an input with is by testing whether the type of an input is compatible with the type of a sub-template. If so, the sub-template is listed in the combo-box for the input in the BEASTObjectInputEditor. The mainid id identifies the BEASTObject that has the type listed in class. Note the $(n) part of the mainid attribute. A sub-template is created in the context of a partition, and wherever there is the string $(n) in the sub-template this is replaced by the name of the partition.
The sub-template starts with a CDATA-section describing the model. The order is not important, but here it was chosen to start with the five parameters that can be estimated for the model, the birth and death rates, interval times, sampling rate and original root height. Then there is an integer parameter that probably should be an integer input for the number of intervals in the skyline model. These parameters are used by the BirthDeathSkylineModel BEASTObject defined below. The BEASTObject has a tree as input, and assumes the main-template provides a tree with id Tree.t:$(n) where $(n) the partition name.
The parameters need a proper prior, defined in the following few lines.
We do not only want to log the parameters, but also a few functions of parameters, namely the sampling rate divided by the sum of death and sampling rate. The RPNcalculator is a BEASTObject that can be used to calculate simple arithmetic expressions in reverse Polish notation, which is useful for logging.
]]>
To complete the sub-template, we need to define rules for connecting the BEASTObjects defined in the CDATA-section with relevant parts of the main template. BeautiConnector specifies a BEASTObject to connect from through the srcID attribute, a BEASTObject to connect to through the targetID template and a inputName specifies the name of the input in the target BEASTObject to connect with. Finally, the connector is only activated when some conditions are met. If the connection is not met, BEAUti will attempt to disconnect the link (if it exists). The conditions are separated by the ‘and’ keyword in the if attribute. The conditions are typically of the form inposterior(BirthDeathSky.$(n)), which tests whether the BEASTObject with id BirthDeathSky.$(n) is a predecessor of a BEASTObject with id posterior in the model. Further, there are conditions of the form Tree.$(n)/estimate=true, are typically used to test whether an input value of a BEASTObject is a certain value. This is mostly relevant to test whether StateNodes are estimated or not, since if they are not estimated no operator should be defined on it, and logging is not very useful. Let’s have a look at a few connectors.
Connectors are tested in order of appearance. It is always a good idea to make the first connector the one connecting the main BEASTObject in the sub-template, since if this main BEASTObject is disconnected, most of the others should be disconnected as well. For this tree-prior, the tree’s estimate flag can become false when the tree for the partition is linked.
end{lstlisting}}
Next, connect priors on parameters to the prior BEASTObject provided by the main template.
{color{blue}scriptsizebegin{lstlisting}[language=XML,firstnumber=last,breaklines=true]
Connect scale operators, and up-down operator. Note that the up-down operator takes as input the birth, death and sampling rate, but if one of these rates is fixed by the user in BEAUti, the link should be disconnected. So, there are three rules for deciding whether these rates should be connected to the operator.