30 June 2015 by Remco Bouckaert
The unwelcome answer to that question is; it depends.
Number of lineages per species
First of all, there should be more than one lineage (= one haploid sequence) for every species. If there is only a single lineage, there are no coalescent events possible in the branches ending in the tips, and the branch above it will have on average only a single coalescent event. This means that the populations sizes for each of the branches will be informed by only a single coalescent event (on average) and there will be very little signal to inform population sizes. The result is that almost certainly, the population size will be sampled from the prior. And since population size and branch lengths are confounded (large population size means larger branch lengths) and the prior on population sizes is quite broad by default, it may take a lot of time to converge.
So, multiple lineages per species is recommended. Of course, this has to be balanced with the penalty in computational that is incurred. So, you have to experiment a bit to find out what is computationally feasible, and how much signal can be obtained from the data.
Sequence length
In SNAPP, every site in a sequence has its own gene tree that is assumed to be independent of all other gene trees. So, adding sites also means adding gene trees.
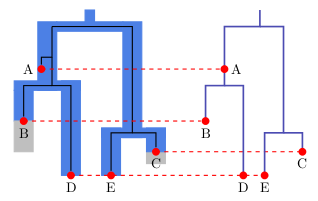
When samples are very closely related, all coalescent events happen very closely to the present time (the sampling time). If so and you look at a branch ending in a species, there is only a single lineage left at the top of the branch. This means we are running in the problem described above; there is no signal in the data left to determine population sizes, and convergence will be difficult. There is no point in adding more sites that have this property, since it would just slow down the calculation without adding more information.
When samples are very distantly related, all coalescent events happen in the branch stemming out of the root. This means, there is no topological information in such samples, and every species tree will fit equally well. On top of this, there is no information to inform population sizes, so SNAPP will not give a lot of information, and will have a terrible time to reach convergence.
In between these extremes, there is the goldilocks zone, where samples coalesc not too early, and not too late, but just in at the right time. In this goldilocks zone, there will be some lineage sorting, so branches above those ending in tips will contain some population size information. This is the kind of data you would like to add.
Of course, it is hard to tell beforehand what kind of data you have, so it is hard to tell beforehand what is the ideal sequence length.